Searches the PDB for candidate protein structural alignments #Search PDB #Find alignment #Sequence alignment #Search #PDB #Aligment
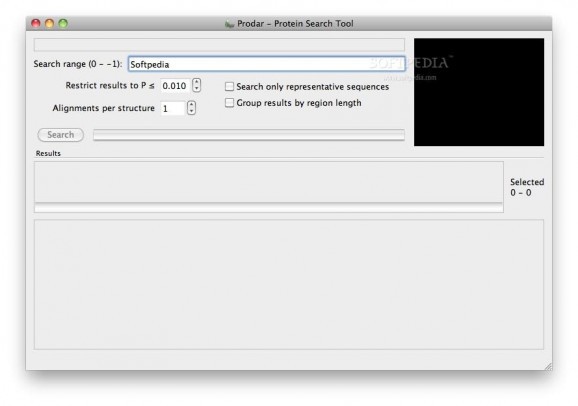
Prodar is an easy to use search application that queries the PDB for candidate structural alignments.
The input to the search is a protein backbone structure read from a standard (text) PDB file, and the results returned are based solely on structural similarity of the backbone without any regard to sequence information. Searches are extremely fast, searching the PDB (included in app) in less than a minute typically.
Prodar identifies partial matches, such that a relatively small section of the query structure may be matched against sections of other structures in the PDB regardless of the relative size of the chains.
The intention is that the tool automatically identifies common domains and motifs in otherwise dissimilar protein structures. These can then be structurally aligned in a different tool to find an RMSD (Prodar just makes suggestions).
Prodar encourages an interactive style of discovery, where searches are rapidly refined and rerun. The results can be surprising!
A detailed guide on how to use the Prodar utility on your Mac is available HERE.
Prodar 1.5
add to watchlist add to download basket send us an update REPORT- runs on:
- Mac OS X (PPC & Intel)
- file size:
- 732 MB
- main category:
- Math/Scientific
- developer:
- visit homepage
Windows Sandbox Launcher
ShareX
Context Menu Manager
calibre
4k Video Downloader
7-Zip
Zoom Client
IrfanView
Microsoft Teams
Bitdefender Antivirus Free
- IrfanView
- Microsoft Teams
- Bitdefender Antivirus Free
- Windows Sandbox Launcher
- ShareX
- Context Menu Manager
- calibre
- 4k Video Downloader
- 7-Zip
- Zoom Client