A streamlined open-source scientific application for all major platforms designed to help you create and curate genome-scale metabolic networks. #Metabolic network creator #Curate metabolic network #Annotated reconstruction #Genome #Cheminformatics #Metabolism
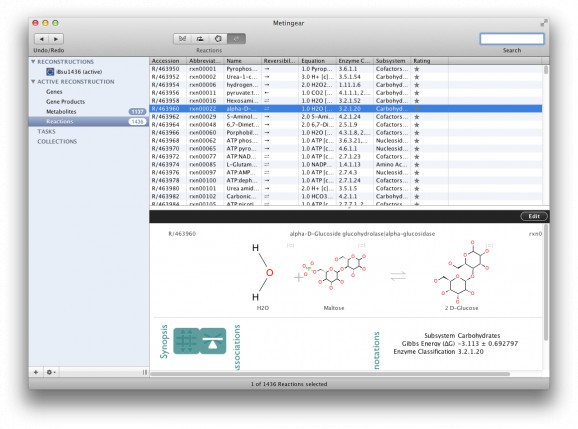
Metingear is a free and cross-platform software solution that enables you to produce and curate metabolic networks with chemical structure on a genomic scale.
Metingear was developed because the process of creating a high-quality genome-scale metabolite reconstruction is not an easy task and might take endless hours of manual work.
Thus, Metingear aims to provide a better and more efficient method for annotated reconstructions of metabolic networks. No need for manual annotation.
You will be able to import existing reconstructions from Microsoft Excel, KEGG Markup Language (KGML) and Systems Biology Markup Language (SBML).
Metingear also offers the ability to include protein and nucleotide sequence information from FASTA format files as well as European Nucleotide Archive (ENA).
Metingear is cross-platform and it works on Mac OS X, Windows and Linux. Binaries for the Windows and Linux platforms are available on the project's homepage.
Metingear 0.9.1 Beta
add to watchlist add to download basket send us an update REPORT- runs on:
- Mac OS X (PPC & Intel)
- file size:
- 34.8 MB
- filename:
- Metingear-0.9.1.dmg
- main category:
- Math/Scientific
- developer:
- visit homepage
Bitdefender Antivirus Free
calibre
Windows Sandbox Launcher
Context Menu Manager
Zoom Client
7-Zip
Microsoft Teams
4k Video Downloader
ShareX
IrfanView
- 4k Video Downloader
- ShareX
- IrfanView
- Bitdefender Antivirus Free
- calibre
- Windows Sandbox Launcher
- Context Menu Manager
- Zoom Client
- 7-Zip
- Microsoft Teams