Likelihood analysis with metropolis algorithm using random coalescence #Estimate recombination rate #Analyze migration rate #Population size estimator #Estimate #Population size #Recombination rate
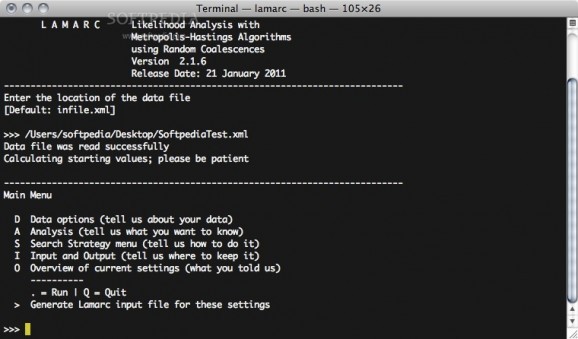
LAMARC is an open source and free command line based tool which estimates population-genetic parameters such as population size, recombination rate, population growth rate, and migration rates.
Moreover, LAMARC is a utility for doing Likelihood Analysis with Metropolis Algorithm using Random Coalescence.
LAMARC estimates effective population sizes, population exponential growth rates, a recombination rate, and past migration rates for one to n populations assuming a migration matrix model with asymmetric migration rates and different subpopulation sizes.
LAMARC can use DNA or RNA sequence data, SNPs, microsatellites, or electrophoretic data.
The LAMARC program can produce: estimates of recombination rate, migration rates between each population pair, population sizes (assuming constant mutation rates among loci), population exponential growth rates, profile likelihood tables, and percentiles.
What's new in LAMARC 2.1.10:
- FIXED: lam_conv file converter reversed source and destination populations when using custom migration matrices. If you used a custom matrix to set asymmetrical migration rates in any prior version, you will need to re-convert your files and re-run your analysis. All other results are unaffected.
LAMARC 2.1.10
add to watchlist add to download basket send us an update REPORT- runs on:
- Mac OS X (-)
- file size:
- 5.6 MB
- filename:
- lamarc-2.1.10-osx.dmg
- main category:
- Math/Scientific
- developer:
- visit homepage
IrfanView
Bitdefender Antivirus Free
4k Video Downloader
calibre
7-Zip
Zoom Client
Context Menu Manager
ShareX
Windows Sandbox Launcher
Microsoft Teams
- ShareX
- Windows Sandbox Launcher
- Microsoft Teams
- IrfanView
- Bitdefender Antivirus Free
- 4k Video Downloader
- calibre
- 7-Zip
- Zoom Client
- Context Menu Manager