A free and open-source general purpose multiple sequence alignment program that enables you to perform profile alignments or phylogenetic trees. #Align sequence #Dna sequence alignment #Sequence match calculator #Calculate #Align #Alignment
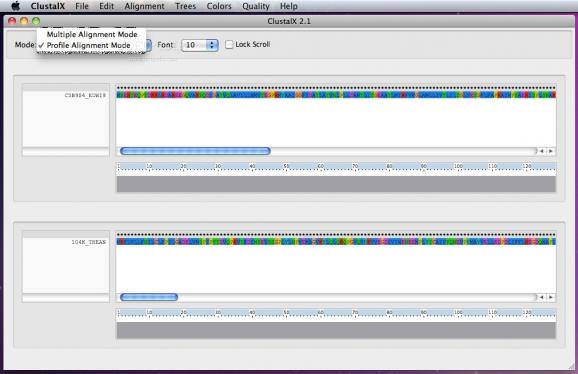
ClustalX is a streamlined OS X utility that provides the necessary tools to align DNA or protein sequences from within a user-friendly interface or a Terminal window.
ClustalX comes with support for numerous input formats, such as GDE, FASTA, NBRF/PIR, GCG9 RSF, Clustal, GCC/MSF and EMBL/Swiss-Prot.
In order to use ClustalX, you have to place each sequence in one file and arrange them one after another.
ClustalX's intuitive interface enables you to perform profile alignments, phylogenetic trees and multiple alignment in just a few easy steps.
From the Edit menu you can easily search for a String, remove gaps, clear sequences or range selections and switch between profiles.
By accessing the Alignment menu, you can easily align the sequences to the desired profile and open the Output Format Option window.
ClustalX also allows you to export to the NEXUS, Clustral, GDE, GCG/MSF and NBRF/PIR format.
ClustalX enables its users to choose between existing color sets or load a color parameter file with just a mouse click.
On top of that, users who are more comfortable performing all aforementioned tasks from a Terminal window, can download and use ClustalW.
Users that are interested in more advanced features like using secondary structures for profile alignment can use the "Using ClustalX for multiple sequence" tutorial created by by Jarno Tuimala.
What's new in ClustalX 2.1:
- Fixed bug 196 "clustalx: user feedback about use of secondary structure printed to console" - secondary structure is now used if specified in Alignment -> Alignment Parameters -> Secondary Structure Parameters UserParameters->getGui() should be used when ClustalW code needs to know if a function has been called from ClustalX
- Fixed bug 204 "Nexus alignment format contain invalid line" - the amino acid alphabet line has been removed
- Missing/corrupted file names in ClustalX status messages have been fixed
- Fixed bug 175 "msf/pileup files cannot be read if sequences names are all numbers" - this happened if a line such as 528244 .......... .......... .......... .......... .......... was present in the first block of the file
ClustalX 2.1
add to watchlist add to download basket send us an update REPORT- runs on:
- Mac OS X (PPC & Intel)
- file size:
- 12.6 MB
- filename:
- clustalx-2.1-macosx.dmg
- main category:
- Math/Scientific
- developer:
- visit homepage
IrfanView
Windows Sandbox Launcher
7-Zip
Bitdefender Antivirus Free
Zoom Client
Microsoft Teams
calibre
4k Video Downloader
Context Menu Manager
ShareX
- 4k Video Downloader
- Context Menu Manager
- ShareX
- IrfanView
- Windows Sandbox Launcher
- 7-Zip
- Bitdefender Antivirus Free
- Zoom Client
- Microsoft Teams
- calibre