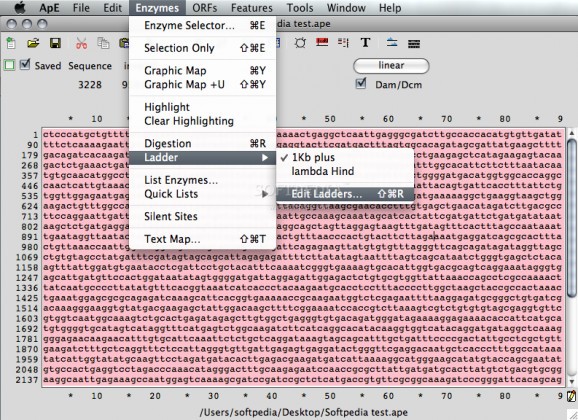
An easy to use plasmid editor for your Mac. #Plasmid editor #Plasmid analysis #Dna analyzer #Plasmid #Dna #Editor
ApE 2.0.47
add to watchlist add to download basket send us an update REPORT- runs on:
- Mac OS X 10.5 or later (Intel only)
- file size:
- 7.6 MB
- filename:
- Ape_OSX_current.dmg
7 screenshots:
- main category:
- Math/Scientific
- developer:
- visit homepage
ShareX
Capture your screen, create GIFs, and record videos through this versatile solution that includes various other amenities: an OCR scanner, image uploader, URL shortener, and much more
4k Video Downloader
Export your favorite YouTube videos and playlists with this intuitive, lightweight program, built to facilitate downloading clips from the popular website
calibre
Effortlessly keep your e-book library thoroughly organized with the help of the numerous features offered by this efficient and capable manager
Zoom Client
The official desktop client for Zoom, the popular video conferencing and collaboration tool used by millions of people worldwide
Windows Sandbox Launcher
Set up the Windows Sandbox parameters to your specific requirements, with this dedicated launcher that features advanced parametrization
Microsoft Teams
Effortlessly chat, collaborate on projects, and transfer files within a business-like environment by employing this Microsoft-vetted application
IrfanView
With support for a long list of plugins, this minimalistic utility helps you view images, as well as edit and convert them using a built-in batch mode
Context Menu Manager
Customize Windows’ original right-click context menu using this free, portable and open-source utility meant to enhance your workflow
Bitdefender Antivirus Free
Feather-light and free antivirus solution from renowned developer that keeps the PC protected at all times from malware without requiring user configuration
7-Zip
An intuitive application with a very good compression ratio that can help you not only create and extract archives, but also test them for errors
% discount
Context Menu Manager
- Context Menu Manager
- Bitdefender Antivirus Free
- 7-Zip
- ShareX
- 4k Video Downloader
- calibre
- Zoom Client
- Windows Sandbox Launcher
- Microsoft Teams
- IrfanView
essentials
Click to load comments
This enables Disqus, Inc. to process some of your data. Disqus privacy policy